#load required packages
library(tidyverse)
library(tidymodels)
library(here)
library(rpart)
library(glmnet)
library(ranger)
library(rpart.plot)
library(parallel)
library(vip)Flu Analysis
Machine Learning
This file contains data processing and analysis, including some implementation of machine learning. It is conducted on the dataset from McKay et al 2020, found here.
Load and Process Data
#load data
flu_data <- readr::read_rds(here("fluanalysis", "data", "flu_data_clean.RDS"))
glimpse(flu_data)Rows: 730
Columns: 32
$ SwollenLymphNodes <fct> Yes, Yes, Yes, Yes, Yes, No, No, No, Yes, No, Yes, Y~
$ ChestCongestion <fct> No, Yes, Yes, Yes, No, No, No, Yes, Yes, Yes, Yes, Y~
$ ChillsSweats <fct> No, No, Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, Yes, ~
$ NasalCongestion <fct> No, Yes, Yes, Yes, No, No, No, Yes, Yes, Yes, Yes, Y~
$ CoughYN <fct> Yes, Yes, No, Yes, No, Yes, Yes, Yes, Yes, Yes, No, ~
$ Sneeze <fct> No, No, Yes, Yes, No, Yes, No, Yes, No, No, No, No, ~
$ Fatigue <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Ye~
$ SubjectiveFever <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, Yes~
$ Headache <fct> Yes, Yes, Yes, Yes, Yes, Yes, No, Yes, Yes, Yes, Yes~
$ Weakness <fct> Mild, Severe, Severe, Severe, Moderate, Moderate, Mi~
$ WeaknessYN <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Ye~
$ CoughIntensity <fct> Severe, Severe, Mild, Moderate, None, Moderate, Seve~
$ CoughYN2 <fct> Yes, Yes, Yes, Yes, No, Yes, Yes, Yes, Yes, Yes, Yes~
$ Myalgia <fct> Mild, Severe, Severe, Severe, Mild, Moderate, Mild, ~
$ MyalgiaYN <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Ye~
$ RunnyNose <fct> No, No, Yes, Yes, No, No, Yes, Yes, Yes, Yes, No, No~
$ AbPain <fct> No, No, Yes, No, No, No, No, No, No, No, Yes, Yes, N~
$ ChestPain <fct> No, No, Yes, No, No, Yes, Yes, No, No, No, No, Yes, ~
$ Diarrhea <fct> No, No, No, No, No, Yes, No, No, No, No, No, No, No,~
$ EyePn <fct> No, No, No, No, Yes, No, No, No, No, No, Yes, No, Ye~
$ Insomnia <fct> No, No, Yes, Yes, Yes, No, No, Yes, Yes, Yes, Yes, Y~
$ ItchyEye <fct> No, No, No, No, No, No, No, No, No, No, No, No, Yes,~
$ Nausea <fct> No, No, Yes, Yes, Yes, Yes, No, No, Yes, Yes, Yes, Y~
$ EarPn <fct> No, Yes, No, Yes, No, No, No, No, No, No, No, Yes, Y~
$ Hearing <fct> No, Yes, No, No, No, No, No, No, No, No, No, No, No,~
$ Pharyngitis <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, No, No, Yes, ~
$ Breathless <fct> No, No, Yes, No, No, Yes, No, No, No, Yes, No, Yes, ~
$ ToothPn <fct> No, No, Yes, No, No, No, No, No, Yes, No, No, Yes, N~
$ Vision <fct> No, No, No, No, No, No, No, No, No, No, No, No, No, ~
$ Vomit <fct> No, No, No, No, No, No, Yes, No, No, No, Yes, Yes, N~
$ Wheeze <fct> No, No, No, Yes, No, Yes, No, No, No, No, No, Yes, N~
$ BodyTemp <dbl> 98.3, 100.4, 100.8, 98.8, 100.5, 98.4, 102.5, 98.4, ~I now want to remove a few correlated variables.
#remove yes/no variables
flu_data <- flu_data %>%
select(-ends_with("YN"))
#remove near-zero variance predictors (hearing and vision)
flu_data <- flu_data %>%
select(-c(Hearing, Vision))
#view new data
glimpse(flu_data)Rows: 730
Columns: 27
$ SwollenLymphNodes <fct> Yes, Yes, Yes, Yes, Yes, No, No, No, Yes, No, Yes, Y~
$ ChestCongestion <fct> No, Yes, Yes, Yes, No, No, No, Yes, Yes, Yes, Yes, Y~
$ ChillsSweats <fct> No, No, Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, Yes, ~
$ NasalCongestion <fct> No, Yes, Yes, Yes, No, No, No, Yes, Yes, Yes, Yes, Y~
$ Sneeze <fct> No, No, Yes, Yes, No, Yes, No, Yes, No, No, No, No, ~
$ Fatigue <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Ye~
$ SubjectiveFever <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, Yes~
$ Headache <fct> Yes, Yes, Yes, Yes, Yes, Yes, No, Yes, Yes, Yes, Yes~
$ Weakness <fct> Mild, Severe, Severe, Severe, Moderate, Moderate, Mi~
$ CoughIntensity <fct> Severe, Severe, Mild, Moderate, None, Moderate, Seve~
$ CoughYN2 <fct> Yes, Yes, Yes, Yes, No, Yes, Yes, Yes, Yes, Yes, Yes~
$ Myalgia <fct> Mild, Severe, Severe, Severe, Mild, Moderate, Mild, ~
$ RunnyNose <fct> No, No, Yes, Yes, No, No, Yes, Yes, Yes, Yes, No, No~
$ AbPain <fct> No, No, Yes, No, No, No, No, No, No, No, Yes, Yes, N~
$ ChestPain <fct> No, No, Yes, No, No, Yes, Yes, No, No, No, No, Yes, ~
$ Diarrhea <fct> No, No, No, No, No, Yes, No, No, No, No, No, No, No,~
$ EyePn <fct> No, No, No, No, Yes, No, No, No, No, No, Yes, No, Ye~
$ Insomnia <fct> No, No, Yes, Yes, Yes, No, No, Yes, Yes, Yes, Yes, Y~
$ ItchyEye <fct> No, No, No, No, No, No, No, No, No, No, No, No, Yes,~
$ Nausea <fct> No, No, Yes, Yes, Yes, Yes, No, No, Yes, Yes, Yes, Y~
$ EarPn <fct> No, Yes, No, Yes, No, No, No, No, No, No, No, Yes, Y~
$ Pharyngitis <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, No, No, Yes, ~
$ Breathless <fct> No, No, Yes, No, No, Yes, No, No, No, Yes, No, Yes, ~
$ ToothPn <fct> No, No, Yes, No, No, No, No, No, Yes, No, No, Yes, N~
$ Vomit <fct> No, No, No, No, No, No, Yes, No, No, No, Yes, Yes, N~
$ Wheeze <fct> No, No, No, Yes, No, Yes, No, No, No, No, No, Yes, N~
$ BodyTemp <dbl> 98.3, 100.4, 100.8, 98.8, 100.5, 98.4, 102.5, 98.4, ~Data preparation
# set seed for reproducible analysis
set.seed(123)
#split data into 70% training, 30% testing groups
flu_split <-initial_split(flu_data, prop = 7/10, strata = BodyTemp)
#New split dataframes
flu_train <- training(flu_split)
flu_test <- testing(flu_split)Cross validation
#5-fold cross-validation, 5 times repeated, stratified by BodyTemp
folds <- vfold_cv(flu_train, v = 5, repeats = 5, strata = BodyTemp)Recipe
#recipe for all predictors
global_recipe <-
recipe(BodyTemp ~ ., data = flu_train) %>%
step_dummy(all_nominal(), -all_outcomes())
#null recipe
null_recipe <-
recipe(BodyTemp ~ 1, data = flu_train)
#set model engine
linear_reg <- linear_reg() %>%
set_engine("lm") %>%
set_mode("regression")
#create workflow
null_workflow <- workflow() %>%
add_model(linear_reg) %>%
add_recipe(null_recipe)
#fit folds data
null_model <- fit_resamples(null_workflow, resamples = folds)
#null model performance
null_performance <- collect_metrics(null_model)
tibble(null_performance)# A tibble: 2 x 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 rmse standard 1.21 25 0.0177 Preprocessor1_Model1
2 rsq standard NaN 0 NA Preprocessor1_Model1Tree Model
#create recipe for tree model
tree_model <- decision_tree(
cost_complexity = tune(),
tree_depth = tune()
) %>%
set_engine("rpart") %>%
set_mode("regression")
#tree workflow
tree_workflow <- workflow() %>%
add_model(tree_model) %>%
add_recipe(global_recipe)
#setup tree tuning grid
tree_grid <- grid_regular(cost_complexity(),
tree_depth(),
levels = 5)
#fit to folds data
tree_fit <- tune_grid(tree_workflow, resamples = folds, grid = tree_grid)
#null model performance
tree_performance <- collect_metrics(tree_fit)
tibble(tree_performance)# A tibble: 50 x 8
cost_complexity tree_depth .metric .estimator mean n std_err .config
<dbl> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
1 0.0000000001 1 rmse standard 1.19 25 0.0181 Prepro~
2 0.0000000001 1 rsq standard 0.0361 25 0.00422 Prepro~
3 0.0000000178 1 rmse standard 1.19 25 0.0181 Prepro~
4 0.0000000178 1 rsq standard 0.0361 25 0.00422 Prepro~
5 0.00000316 1 rmse standard 1.19 25 0.0181 Prepro~
6 0.00000316 1 rsq standard 0.0361 25 0.00422 Prepro~
7 0.000562 1 rmse standard 1.19 25 0.0181 Prepro~
8 0.000562 1 rsq standard 0.0361 25 0.00422 Prepro~
9 0.1 1 rmse standard 1.21 25 0.0177 Prepro~
10 0.1 1 rsq standard NaN 0 NA Prepro~
# i 40 more rowsautoplot(tree_fit)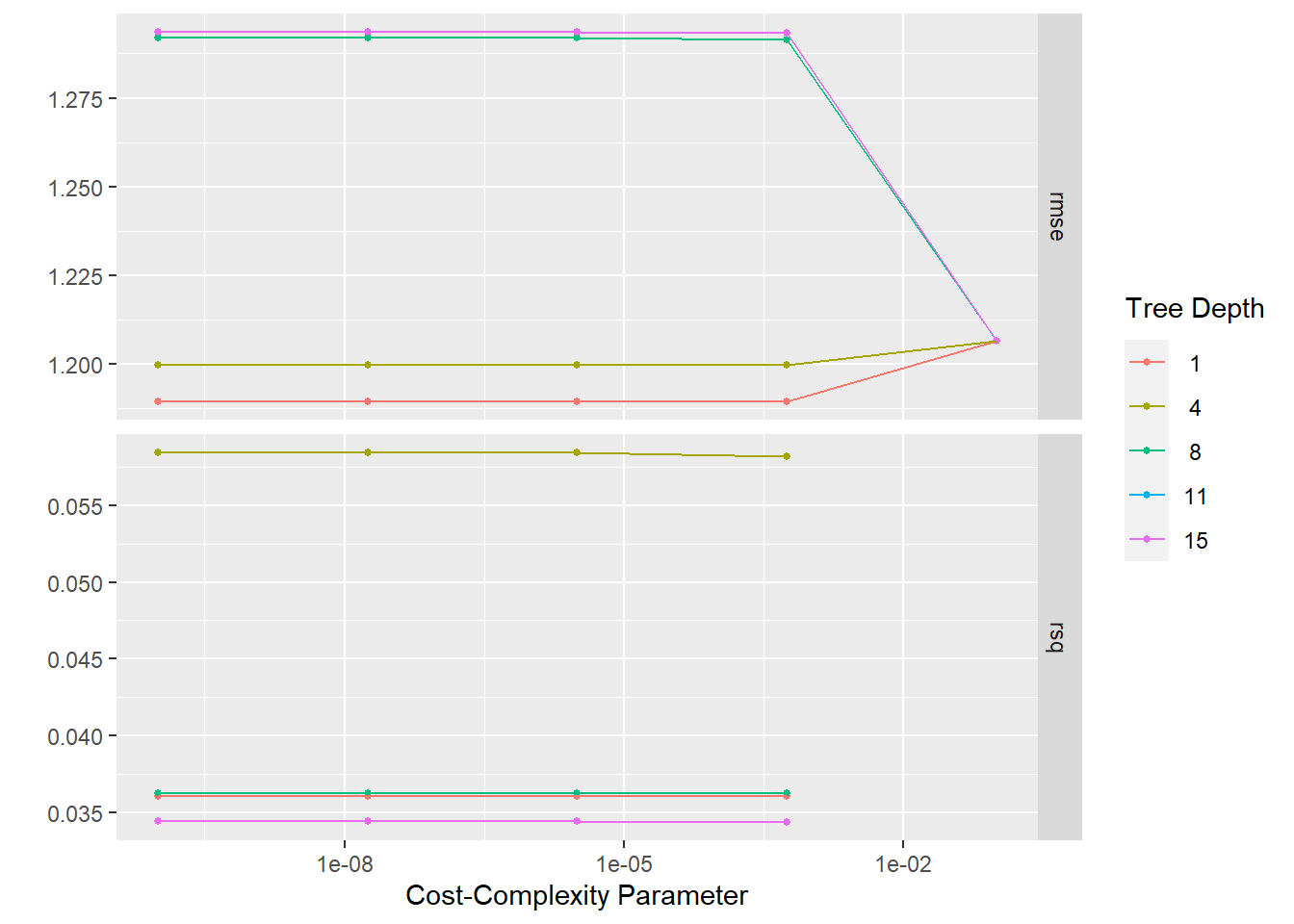
#select best tree model
best_tree <- tree_fit %>%
select_best(metric = 'rmse')
#final model
final_tree <- tree_workflow %>%
finalize_workflow(best_tree)
#fit final model to data
tree_fit_final <- final_tree %>%
fit(flu_train)
#plot final fit
rpart.plot::rpart.plot(extract_fit_parsnip(tree_fit_final)$fit)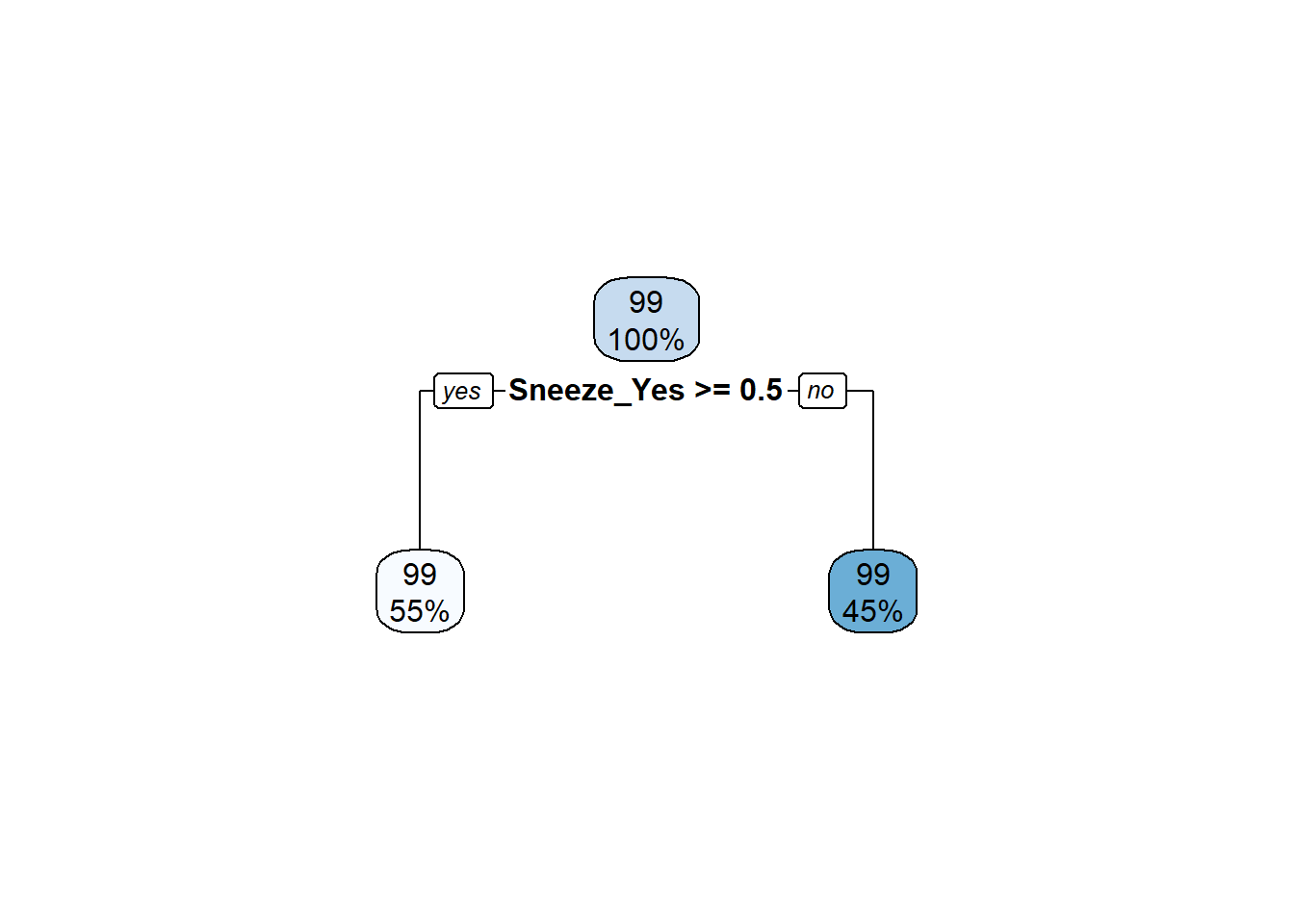
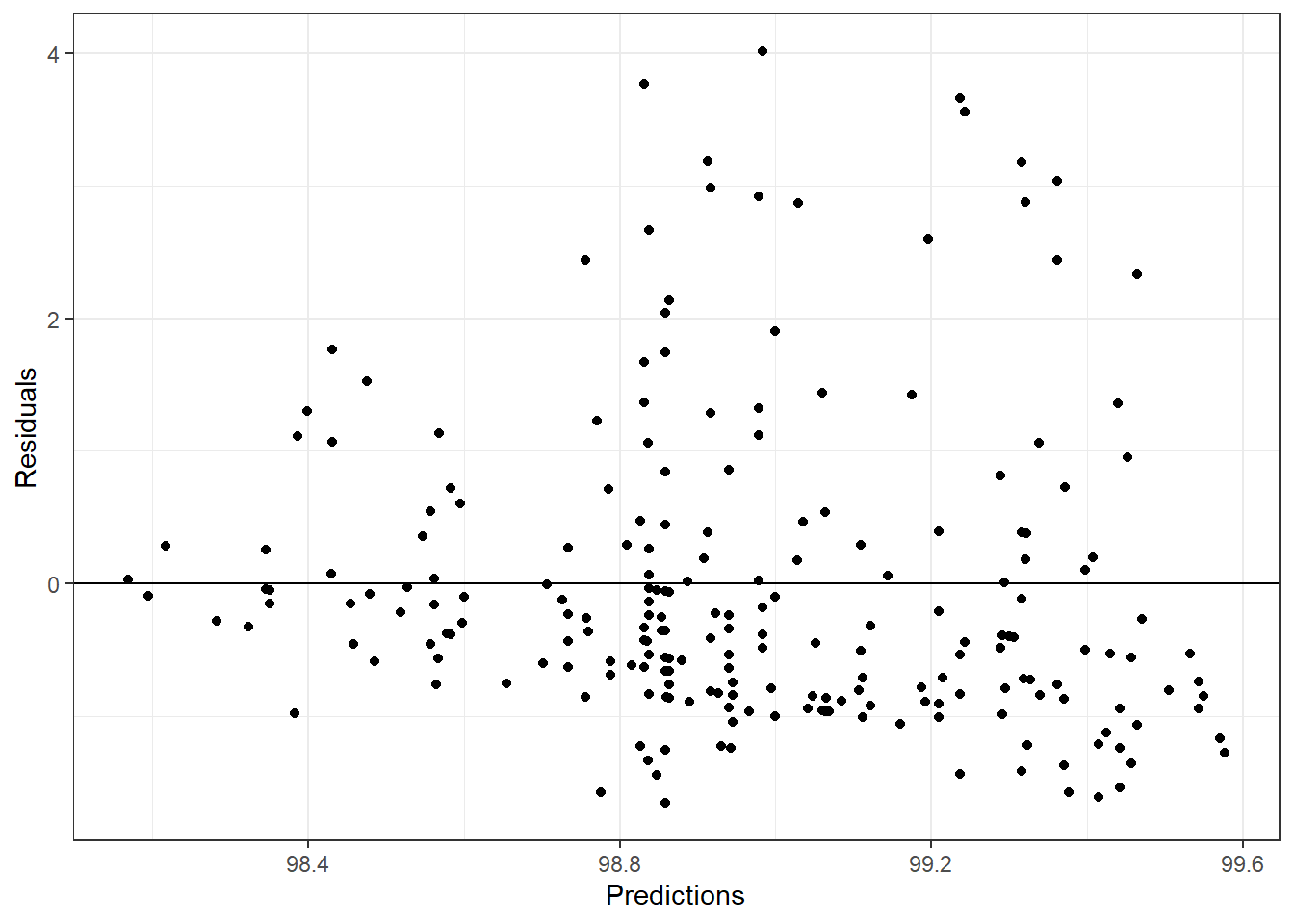
#tree residual plot
tree_pred <- tree_fit_final %>%
fit(flu_train) %>%
predict(flu_train)
tree_resid <- flu_train$BodyTemp - tree_pred$.pred
tree_df <- tibble(tree_resid, tree_pred)
ggplot(aes(.pred, tree_resid), data = tree_df)+
geom_point()+
labs(x = "Predictions", y = "Residuals")+
geom_hline(yintercept = 0)+
theme_bw()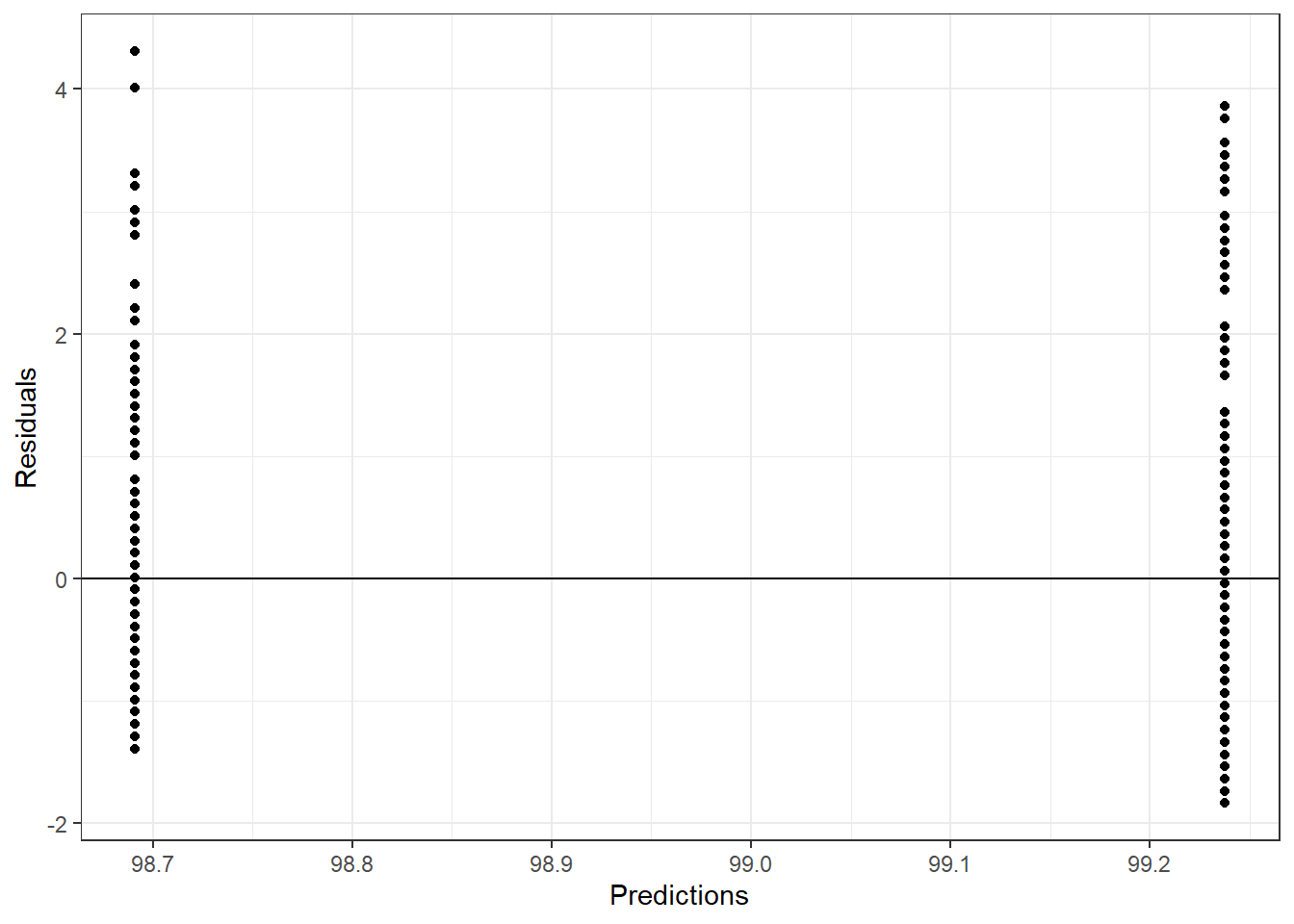
LASSO
#LASSO model
lasso_model <- linear_reg(
penalty=tune(),mixture=1) %>%
set_engine("glmnet") %>%
set_mode("regression")
#LASSO workflow
lasso_workflow <- workflow() %>%
add_model(lasso_model) %>%
add_recipe(global_recipe)
#LASSO grid
lasso_grid <- tibble(penalty = 10^seq(-4, -1, length.out = 30))
#highest and lowest penalty values
lasso_grid %>%
top_n(5)Selecting by penalty# A tibble: 5 x 1
penalty
<dbl>
1 0.0386
2 0.0489
3 0.0621
4 0.0788
5 0.1 lasso_grid %>%
top_n(-5)Selecting by penalty# A tibble: 5 x 1
penalty
<dbl>
1 0.0001
2 0.000127
3 0.000161
4 0.000204
5 0.000259#model tuning and training
lasso_train <- lasso_workflow %>%
tune_grid(
resamples=folds,
grid=lasso_grid,
control=control_grid(save_pred = TRUE),
metrics=metric_set(rmse))
lasso_train %>%
collect_metrics()# A tibble: 30 x 7
penalty .metric .estimator mean n std_err .config
<dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
1 0.0001 rmse standard 1.18 25 0.0167 Preprocessor1_Model01
2 0.000127 rmse standard 1.18 25 0.0167 Preprocessor1_Model02
3 0.000161 rmse standard 1.18 25 0.0167 Preprocessor1_Model03
4 0.000204 rmse standard 1.18 25 0.0167 Preprocessor1_Model04
5 0.000259 rmse standard 1.18 25 0.0167 Preprocessor1_Model05
6 0.000329 rmse standard 1.18 25 0.0167 Preprocessor1_Model06
7 0.000418 rmse standard 1.18 25 0.0167 Preprocessor1_Model07
8 0.000530 rmse standard 1.18 25 0.0167 Preprocessor1_Model08
9 0.000672 rmse standard 1.18 25 0.0167 Preprocessor1_Model09
10 0.000853 rmse standard 1.18 25 0.0167 Preprocessor1_Model10
# i 20 more rowsautoplot(lasso_train)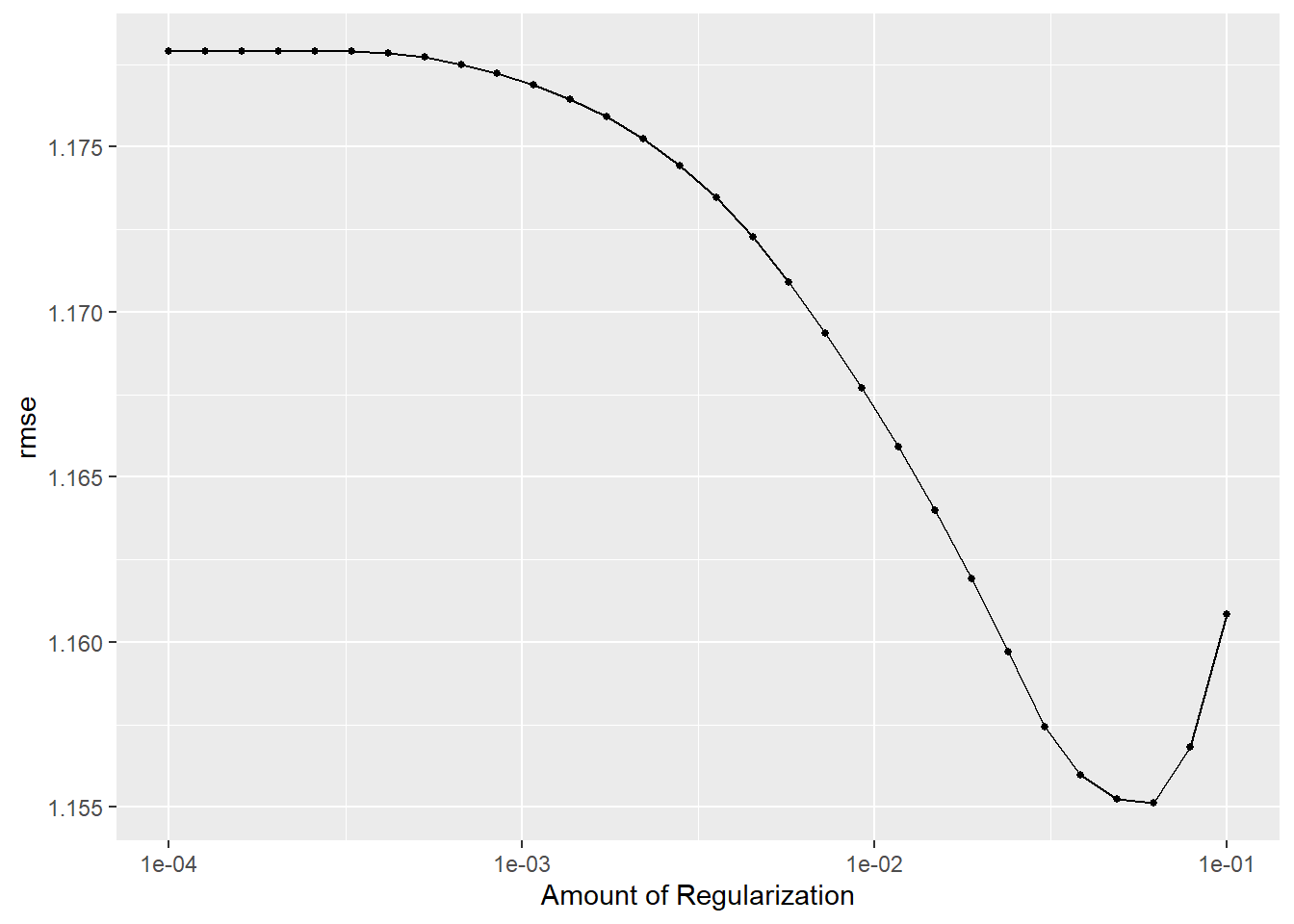
#select best model
lasso_best <- lasso_train %>%
select_best(metric = 'rmse')
#finalized lasso
lasso_final <- lasso_workflow %>%
finalize_workflow(lasso_best)
lasso_fit <- lasso_final %>%
fit(flu_train)
lasso_pred <- lasso_final %>%
fit(flu_train) %>%
predict(flu_train)
plot_fit <- extract_fit_engine(lasso_fit)
plot(plot_fit, "lambda")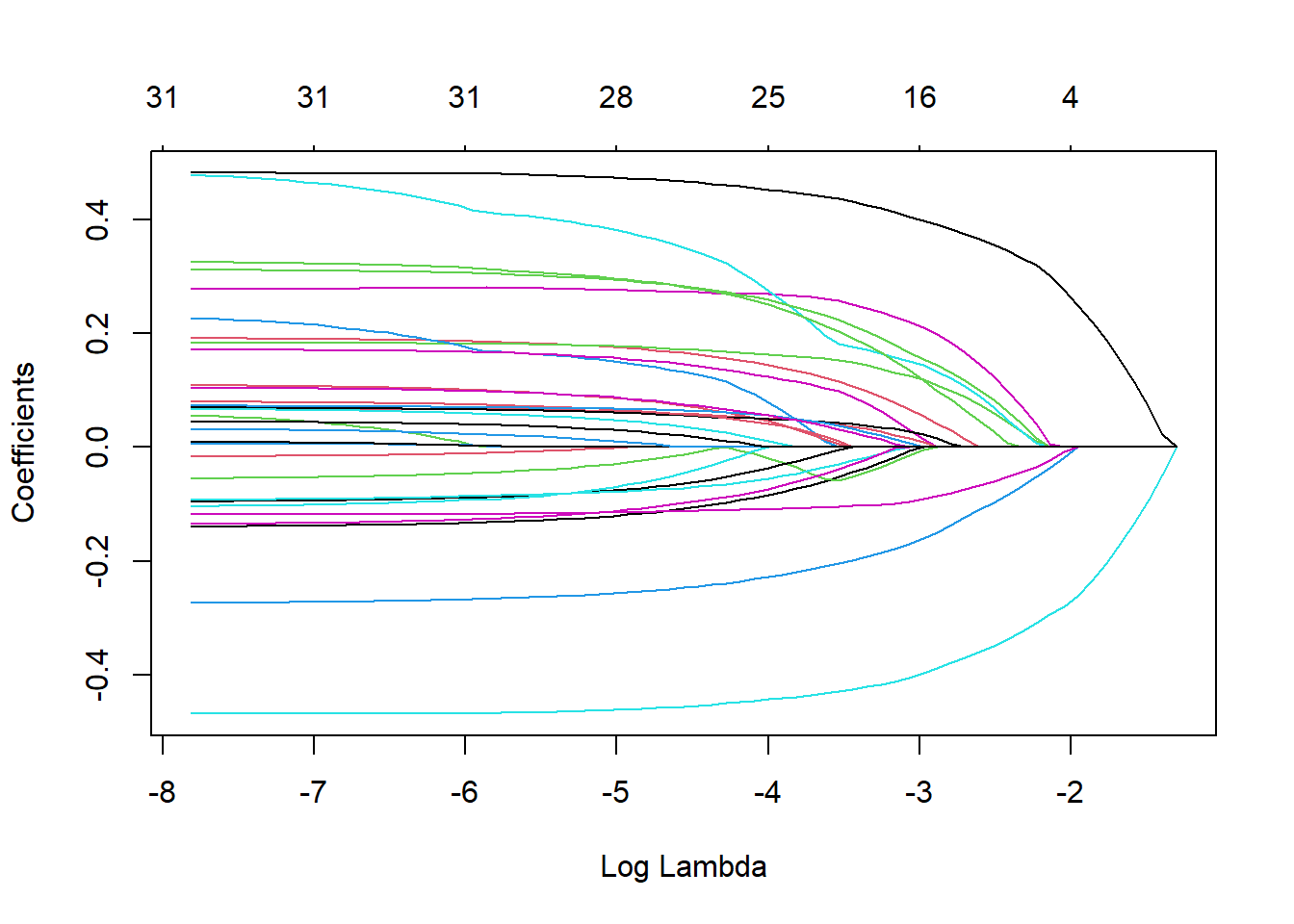
lasso_resid <- flu_train$BodyTemp - lasso_pred$.pred
lasso_df <- tibble(lasso_resid, lasso_pred)
ggplot(aes(.pred, lasso_resid), data = lasso_df)+
geom_point()+
labs(x = "Predictions", y = "Residuals")+
geom_hline(yintercept = 0)+
theme_bw()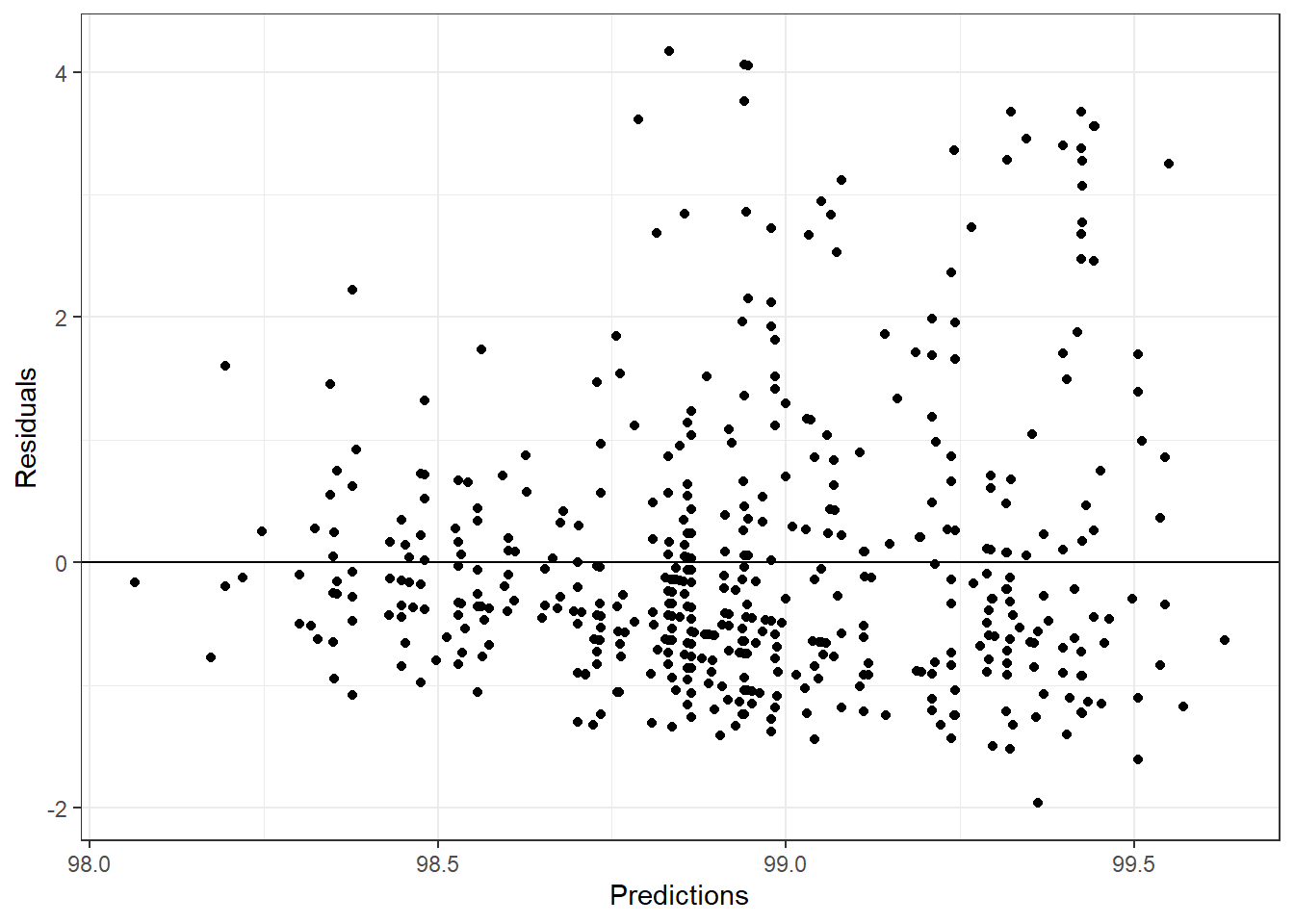
Random Forest
#create cores object
cores <- parallel::detectCores()
#rf model
rf_model <- rand_forest(mtry=tune(),min_n=tune(),trees=1000) %>%
set_engine("ranger", num.threads = cores)%>%
set_mode("regression")
#rf workflow
rf_workflow <- workflow() %>%
add_model(rf_model) %>%
add_recipe(global_recipe)
rf_train <- rf_workflow %>%
tune_grid(
resamples=folds,
grid=25,
control=control_grid(save_pred=TRUE),
metrics = metric_set(rmse))i Creating pre-processing data to finalize unknown parameter: mtryautoplot(rf_train)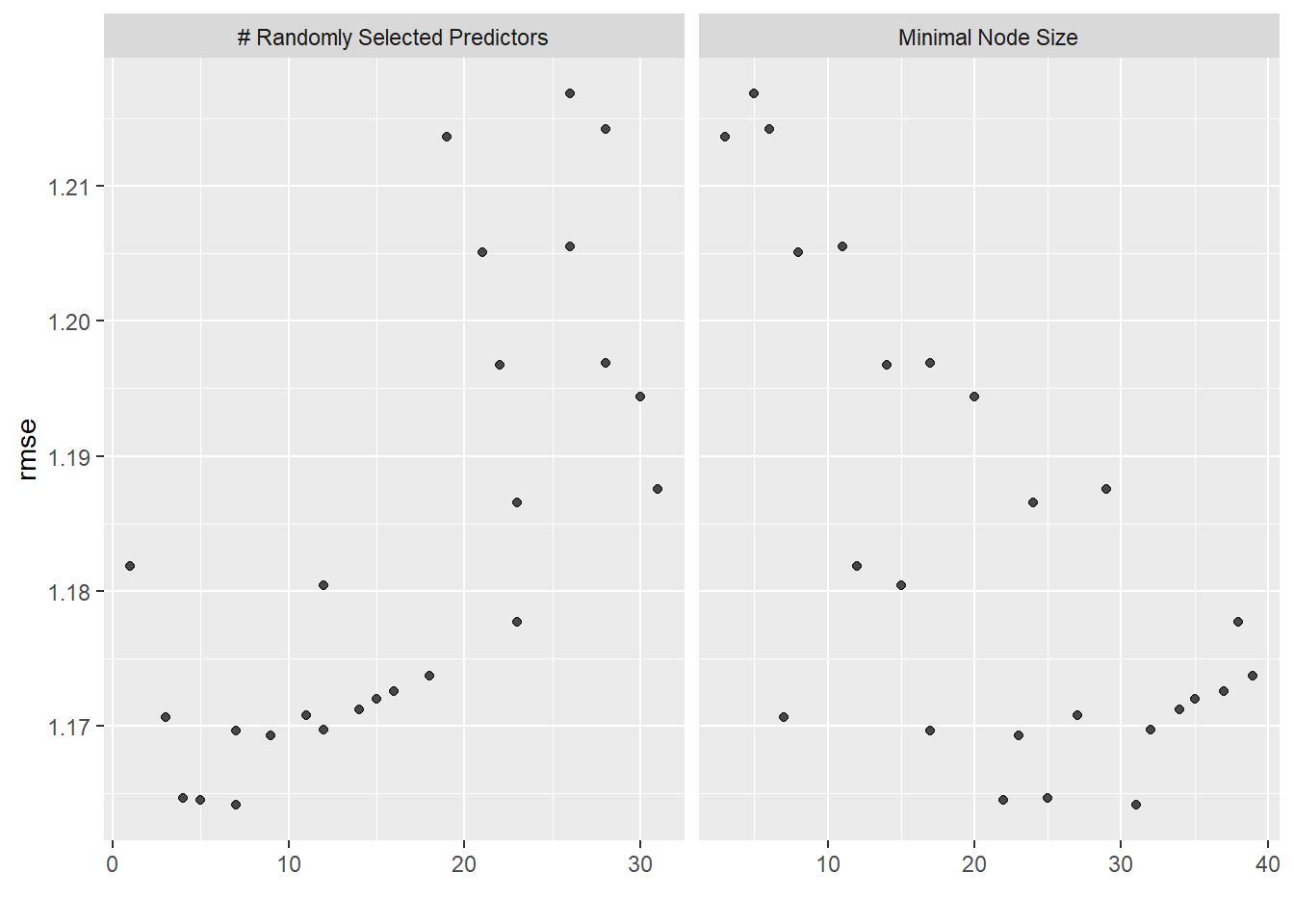
rf_train %>% collect_metrics()# A tibble: 25 x 8
mtry min_n .metric .estimator mean n std_err .config
<int> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
1 30 20 rmse standard 1.19 25 0.0168 Preprocessor1_Model01
2 12 32 rmse standard 1.17 25 0.0166 Preprocessor1_Model02
3 26 11 rmse standard 1.21 25 0.0165 Preprocessor1_Model03
4 28 6 rmse standard 1.21 25 0.0165 Preprocessor1_Model04
5 3 7 rmse standard 1.17 25 0.0166 Preprocessor1_Model05
6 16 37 rmse standard 1.17 25 0.0166 Preprocessor1_Model06
7 4 25 rmse standard 1.16 25 0.0167 Preprocessor1_Model07
8 11 27 rmse standard 1.17 25 0.0168 Preprocessor1_Model08
9 15 35 rmse standard 1.17 25 0.0167 Preprocessor1_Model09
10 23 38 rmse standard 1.18 25 0.0168 Preprocessor1_Model10
# i 15 more rows#select best rf model
rf_best <- rf_train %>%
select_best(metric = 'rmse')
#final rf model
rf_final <- rf_workflow %>%
finalize_workflow(rf_best)
rf_fit <- rf_final %>%
fit(flu_train)
rf_pred <- rf_final %>%
fit(flu_train) %>%
predict(flu_train)
rf_resid <- flu_train$BodyTemp - rf_pred$.pred
rf_df <- tibble(rf_resid, rf_pred)
ggplot(aes(.pred, rf_resid), data = rf_df)+
geom_point()+
labs(x = "Predictions", y = "Residuals")+
geom_hline(yintercept = 0)+
theme_bw()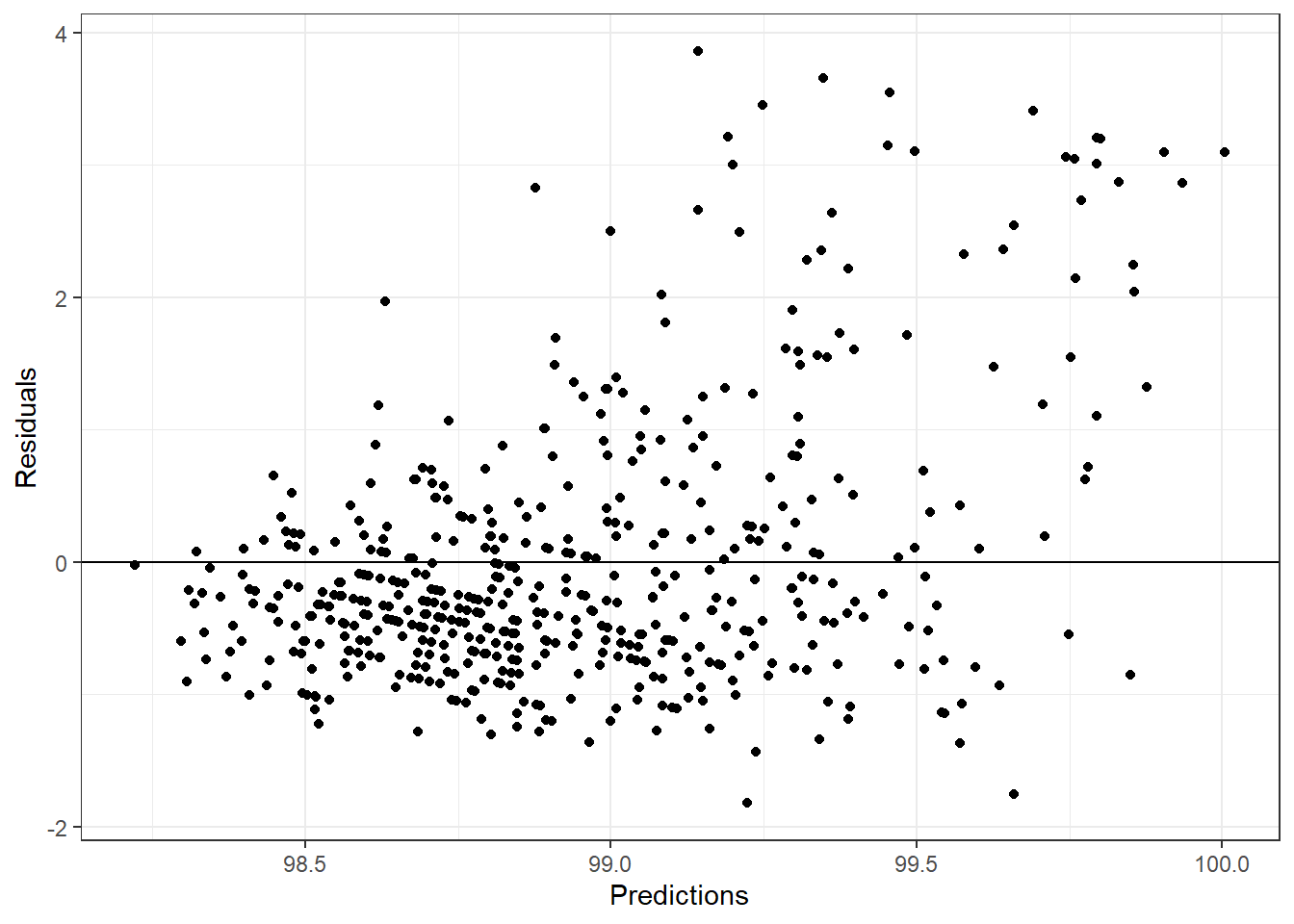
Compare model performances
null_rmse <- null_model %>% show_best(metric = "rmse")
null_rmse # 1.207# A tibble: 1 x 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 rmse standard 1.21 25 0.0177 Preprocessor1_Model1tree_rmse <- tree_fit %>% show_best(metric = "rmse", n=1)
tree_rmse # 1.189# A tibble: 1 x 8
cost_complexity tree_depth .metric .estimator mean n std_err .config
<dbl> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
1 0.0000000001 1 rmse standard 1.19 25 0.0181 Preprocesso~lasso_rmse <- lasso_train %>% show_best(metric = "rmse", n=1)
lasso_rmse # 1.155# A tibble: 1 x 7
penalty .metric .estimator mean n std_err .config
<dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
1 0.0621 rmse standard 1.16 25 0.0170 Preprocessor1_Model28rf_rmse <- rf_train %>% show_best(metric = "rmse", n=1)
rf_rmse # 1.163# A tibble: 1 x 8
mtry min_n .metric .estimator mean n std_err .config
<int> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
1 7 31 rmse standard 1.16 25 0.0165 Preprocessor1_Model15The RMSE values for the best performing models were as follows:
| Null | Tree | LASSO | Random Forest |
|---|---|---|---|
| 1.207 | 1.189 | 1.155 | 1.163 |
Based on these RMSE values, the LASSO appears to have the best performance. Because of this, this model will be used to fit the test data.
final_model <- lasso_final %>%
last_fit(split = flu_split, metrics = metric_set(rmse))
final_model %>% collect_metrics()# A tibble: 1 x 4
.metric .estimator .estimate .config
<chr> <chr> <dbl> <chr>
1 rmse standard 1.15 Preprocessor1_Model1#plot of fit
final_pred <- final_model %>%
collect_predictions()
final_model %>%
extract_fit_engine() %>%
vip()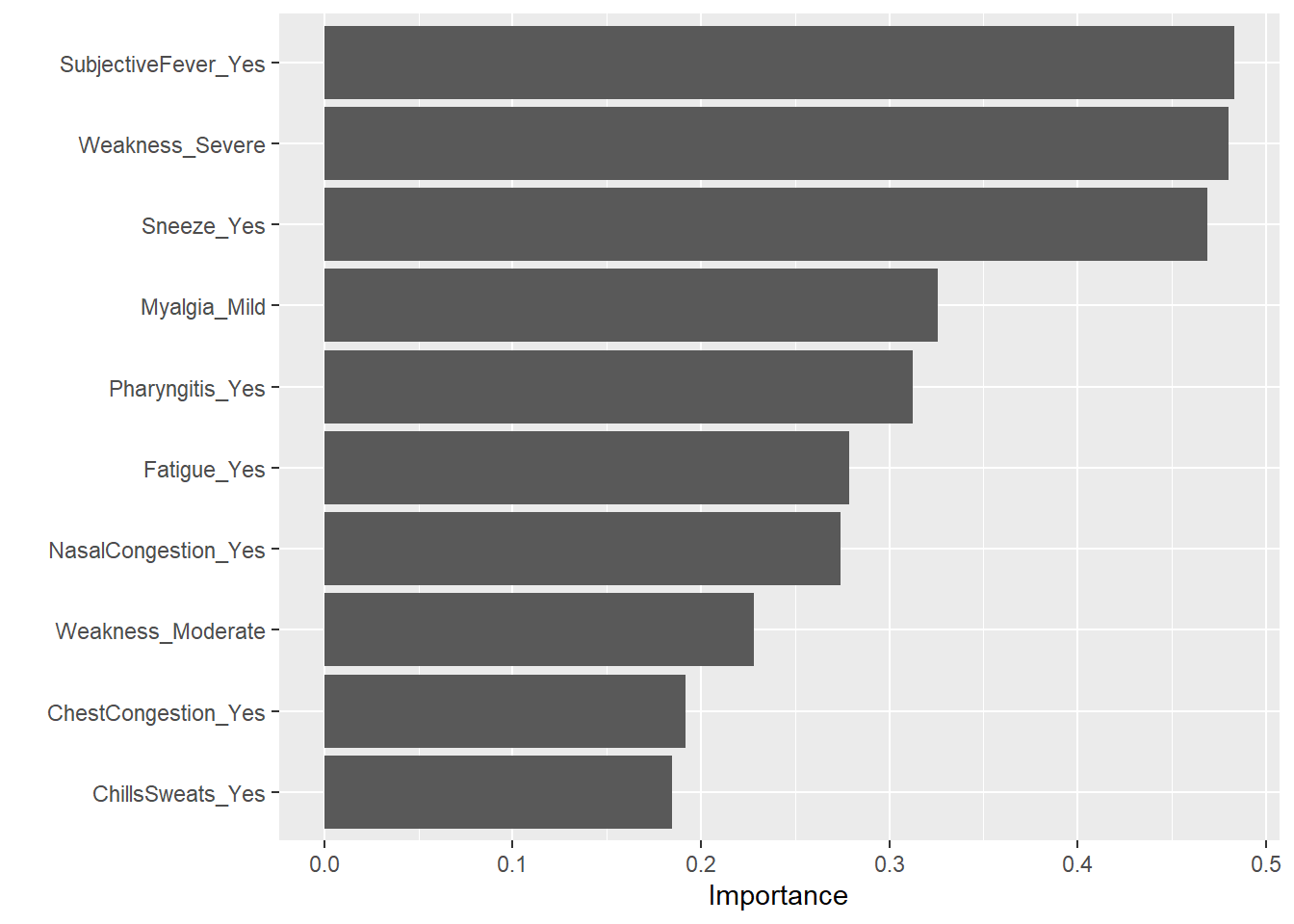
# residual plot
final_resid <- final_pred$BodyTemp - final_pred$.pred
final_df <- tibble(final_resid, final_pred)
ggplot(aes(.pred, final_resid), data = final_df)+
geom_point()+
labs(x = "Predictions", y = "Residuals")+
geom_hline(yintercept = 0)+
theme_bw()