#load necessary packages
library(tidyverse)
library(ggplot2)
library(kableExtra)
library(here)Flu Analysis
Exploration
This is the second file of a four-part data analysis exercise, conducted on the dataset from McKay et al 2020, found here. This file contains data exploration steps in preparation for further analysis.
Load Data/Packages
#load and view data
flu_data <- readRDS(here::here("fluanalysis", "data", "flu_data_clean.RDS"))
glimpse(flu_data)Rows: 730
Columns: 32
$ SwollenLymphNodes <fct> Yes, Yes, Yes, Yes, Yes, No, No, No, Yes, No, Yes, Y~
$ ChestCongestion <fct> No, Yes, Yes, Yes, No, No, No, Yes, Yes, Yes, Yes, Y~
$ ChillsSweats <fct> No, No, Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, Yes, ~
$ NasalCongestion <fct> No, Yes, Yes, Yes, No, No, No, Yes, Yes, Yes, Yes, Y~
$ CoughYN <fct> Yes, Yes, No, Yes, No, Yes, Yes, Yes, Yes, Yes, No, ~
$ Sneeze <fct> No, No, Yes, Yes, No, Yes, No, Yes, No, No, No, No, ~
$ Fatigue <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Ye~
$ SubjectiveFever <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, Yes~
$ Headache <fct> Yes, Yes, Yes, Yes, Yes, Yes, No, Yes, Yes, Yes, Yes~
$ Weakness <fct> Mild, Severe, Severe, Severe, Moderate, Moderate, Mi~
$ WeaknessYN <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Ye~
$ CoughIntensity <fct> Severe, Severe, Mild, Moderate, None, Moderate, Seve~
$ CoughYN2 <fct> Yes, Yes, Yes, Yes, No, Yes, Yes, Yes, Yes, Yes, Yes~
$ Myalgia <fct> Mild, Severe, Severe, Severe, Mild, Moderate, Mild, ~
$ MyalgiaYN <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Ye~
$ RunnyNose <fct> No, No, Yes, Yes, No, No, Yes, Yes, Yes, Yes, No, No~
$ AbPain <fct> No, No, Yes, No, No, No, No, No, No, No, Yes, Yes, N~
$ ChestPain <fct> No, No, Yes, No, No, Yes, Yes, No, No, No, No, Yes, ~
$ Diarrhea <fct> No, No, No, No, No, Yes, No, No, No, No, No, No, No,~
$ EyePn <fct> No, No, No, No, Yes, No, No, No, No, No, Yes, No, Ye~
$ Insomnia <fct> No, No, Yes, Yes, Yes, No, No, Yes, Yes, Yes, Yes, Y~
$ ItchyEye <fct> No, No, No, No, No, No, No, No, No, No, No, No, Yes,~
$ Nausea <fct> No, No, Yes, Yes, Yes, Yes, No, No, Yes, Yes, Yes, Y~
$ EarPn <fct> No, Yes, No, Yes, No, No, No, No, No, No, No, Yes, Y~
$ Hearing <fct> No, Yes, No, No, No, No, No, No, No, No, No, No, No,~
$ Pharyngitis <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, No, No, Yes, ~
$ Breathless <fct> No, No, Yes, No, No, Yes, No, No, No, Yes, No, Yes, ~
$ ToothPn <fct> No, No, Yes, No, No, No, No, No, Yes, No, No, Yes, N~
$ Vision <fct> No, No, No, No, No, No, No, No, No, No, No, No, No, ~
$ Vomit <fct> No, No, No, No, No, No, Yes, No, No, No, Yes, Yes, N~
$ Wheeze <fct> No, No, No, Yes, No, Yes, No, No, No, No, No, Yes, N~
$ BodyTemp <dbl> 98.3, 100.4, 100.8, 98.8, 100.5, 98.4, 102.5, 98.4, ~Explore Data
Let’s take a look at some summary stats for this dataset.
summary(flu_data) SwollenLymphNodes ChestCongestion ChillsSweats NasalCongestion CoughYN
No :418 No :323 No :130 No :167 No : 75
Yes:312 Yes:407 Yes:600 Yes:563 Yes:655
Sneeze Fatigue SubjectiveFever Headache Weakness WeaknessYN
No :339 No : 64 No :230 No :115 None : 49 No : 49
Yes:391 Yes:666 Yes:500 Yes:615 Mild :223 Yes:681
Moderate:338
Severe :120
CoughIntensity CoughYN2 Myalgia MyalgiaYN RunnyNose AbPain
None : 47 No : 47 None : 79 No : 79 No :211 No :639
Mild :154 Yes:683 Mild :213 Yes:651 Yes:519 Yes: 91
Moderate:357 Moderate:325
Severe :172 Severe :113
ChestPain Diarrhea EyePn Insomnia ItchyEye Nausea EarPn
No :497 No :631 No :617 No :315 No :551 No :475 No :568
Yes:233 Yes: 99 Yes:113 Yes:415 Yes:179 Yes:255 Yes:162
Hearing Pharyngitis Breathless ToothPn Vision Vomit Wheeze
No :700 No :119 No :436 No :565 No :711 No :652 No :510
Yes: 30 Yes:611 Yes:294 Yes:165 Yes: 19 Yes: 78 Yes:220
BodyTemp
Min. : 97.20
1st Qu.: 98.20
Median : 98.50
Mean : 98.94
3rd Qu.: 99.30
Max. :103.10 The two main outcomes of interest are BodyTemp and Nausea. I will plot a number of variables against these outcomes to see if any trends are immediately noticeable.
First, I want to look at histogram of body temperature to see the range and distribution.
flu_data %>%
ggplot()+
geom_histogram(aes(BodyTemp))+
theme_classic()`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.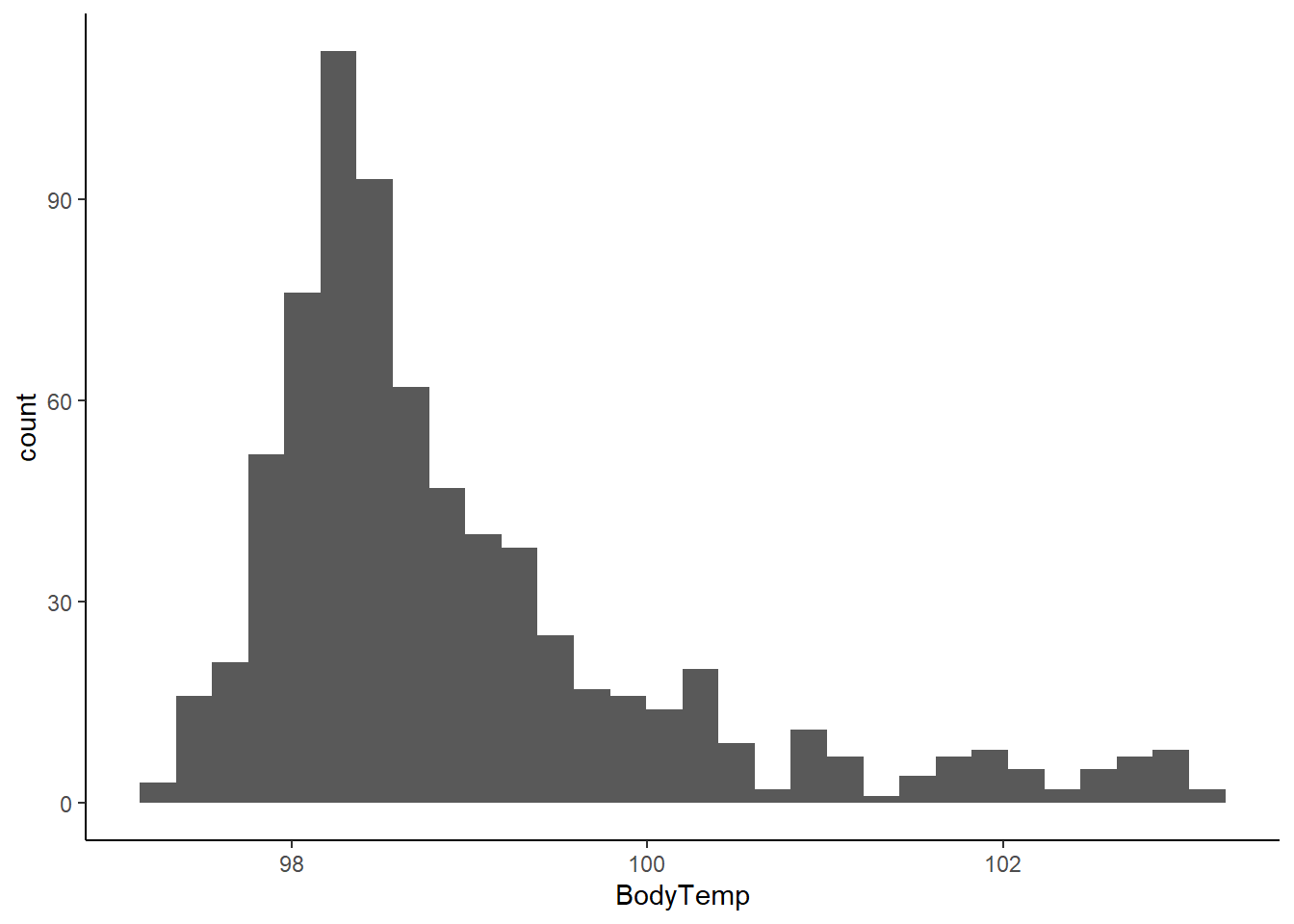
This shows that a majority of patients recorded temperatures in a normal (non-fever) range. A decent proportion had temperatures between 99-100 F, with fewer being above 100 F. Temperatures up to 103.1 F were recorded.
Body Temperature
Now I will look at a number of plots comparing body temperature vs some relevant categorical predictors.
Boxplot of body temperature vs reported fever symptoms.
ggplot(flu_data, aes(BodyTemp, SubjectiveFever))+
geom_boxplot(fill="gray90")+
theme_classic()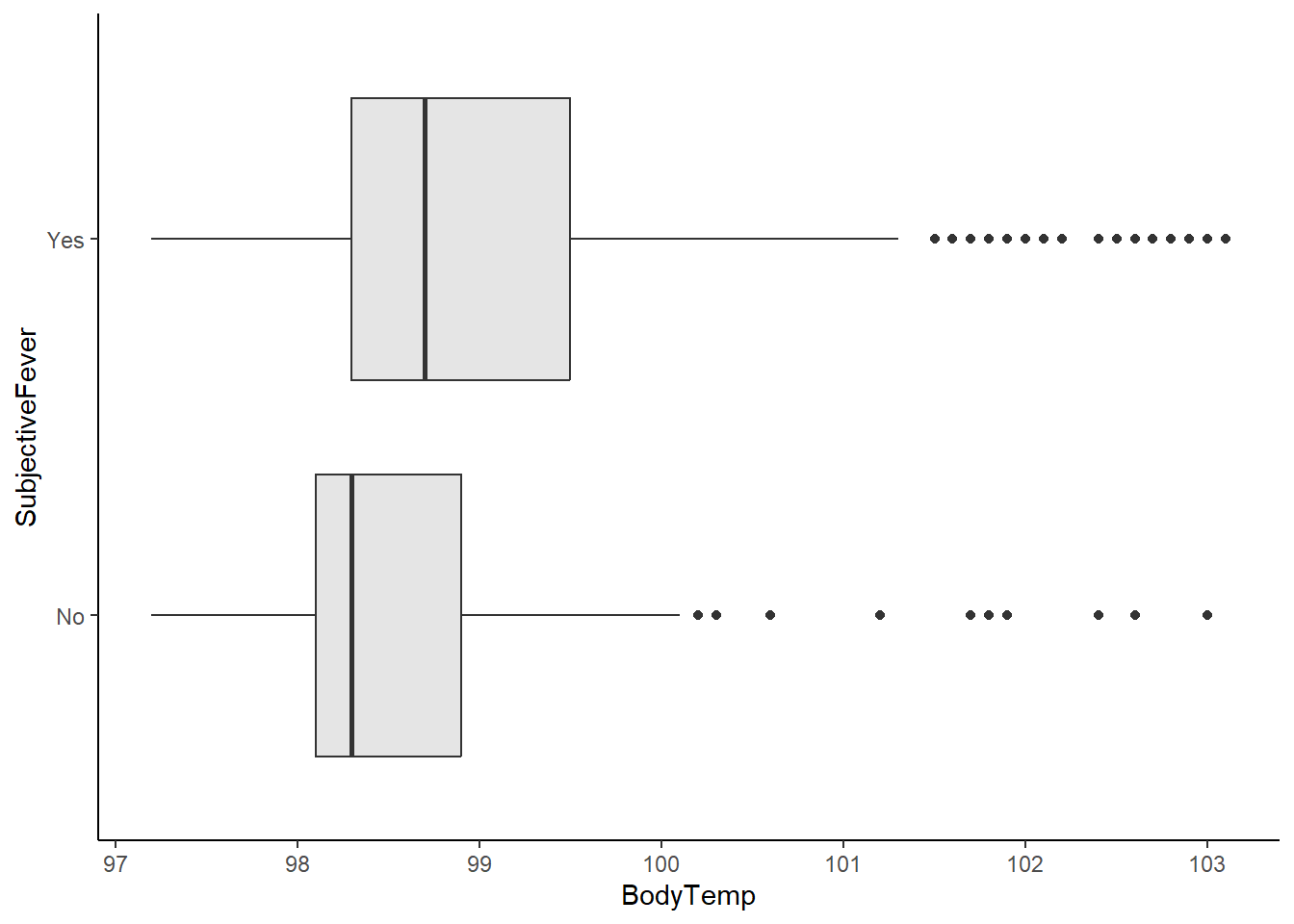
It looks like in general, patients reporting fever symptoms had higher body temperatures than those that did not. The interquartile range for patients reporting fevers falls between normal temperature to low-grade fever. Several patients with body temperatures above 100 F did not report any fever symptoms.
Boxplot of body temperature vs reported chills/sweats.
ggplot(flu_data, aes(BodyTemp, ChillsSweats))+
geom_boxplot(fill="gray90")+
theme_classic()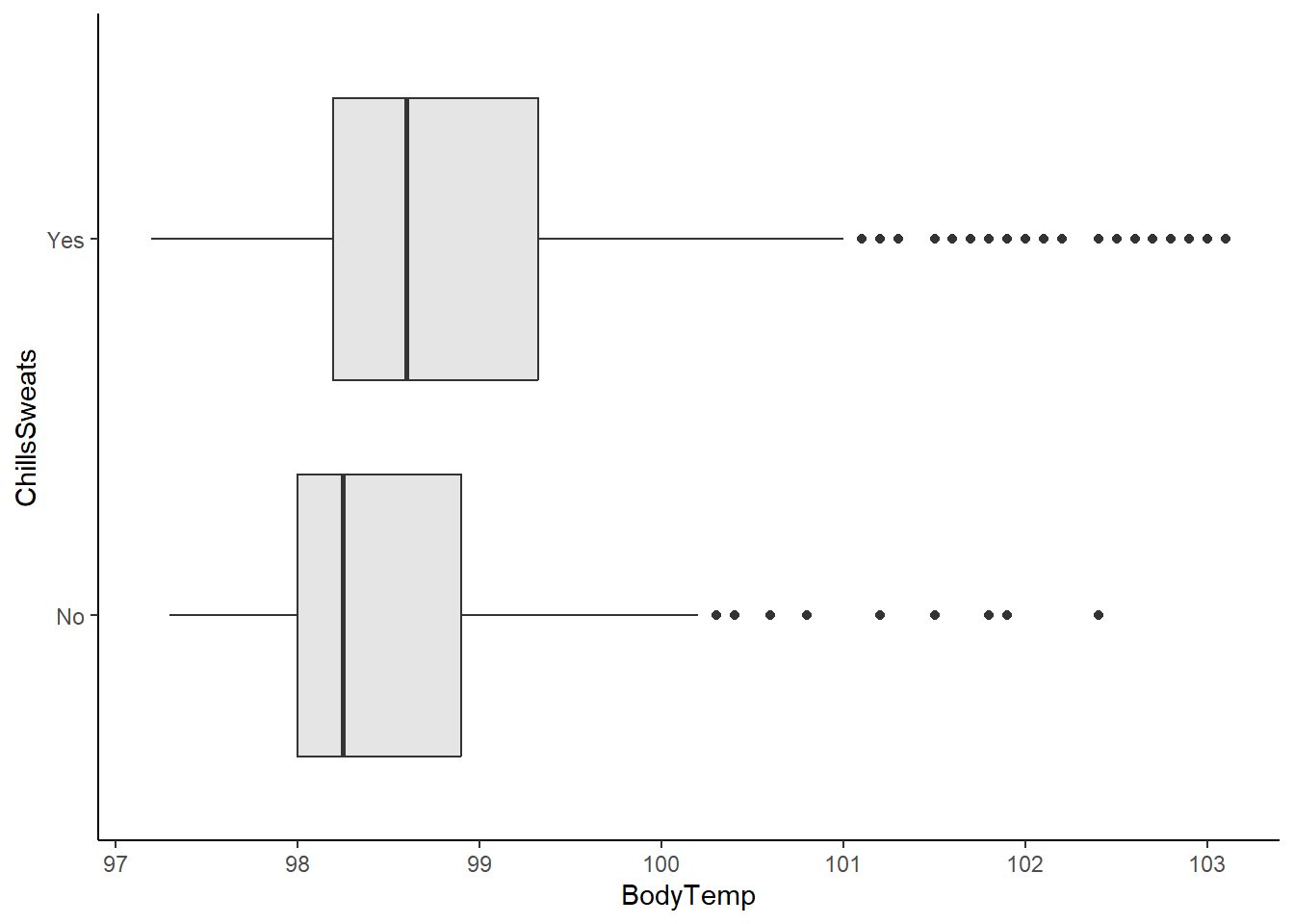
It seems that patients with higher body temperatures experienced chills/sweats slightly more commonly than those with lower body temperatures.
Boxplot of body temperature vs reported weakness.
ggplot(flu_data, aes(BodyTemp, Weakness))+
geom_boxplot(fill="gray90")+
theme_classic()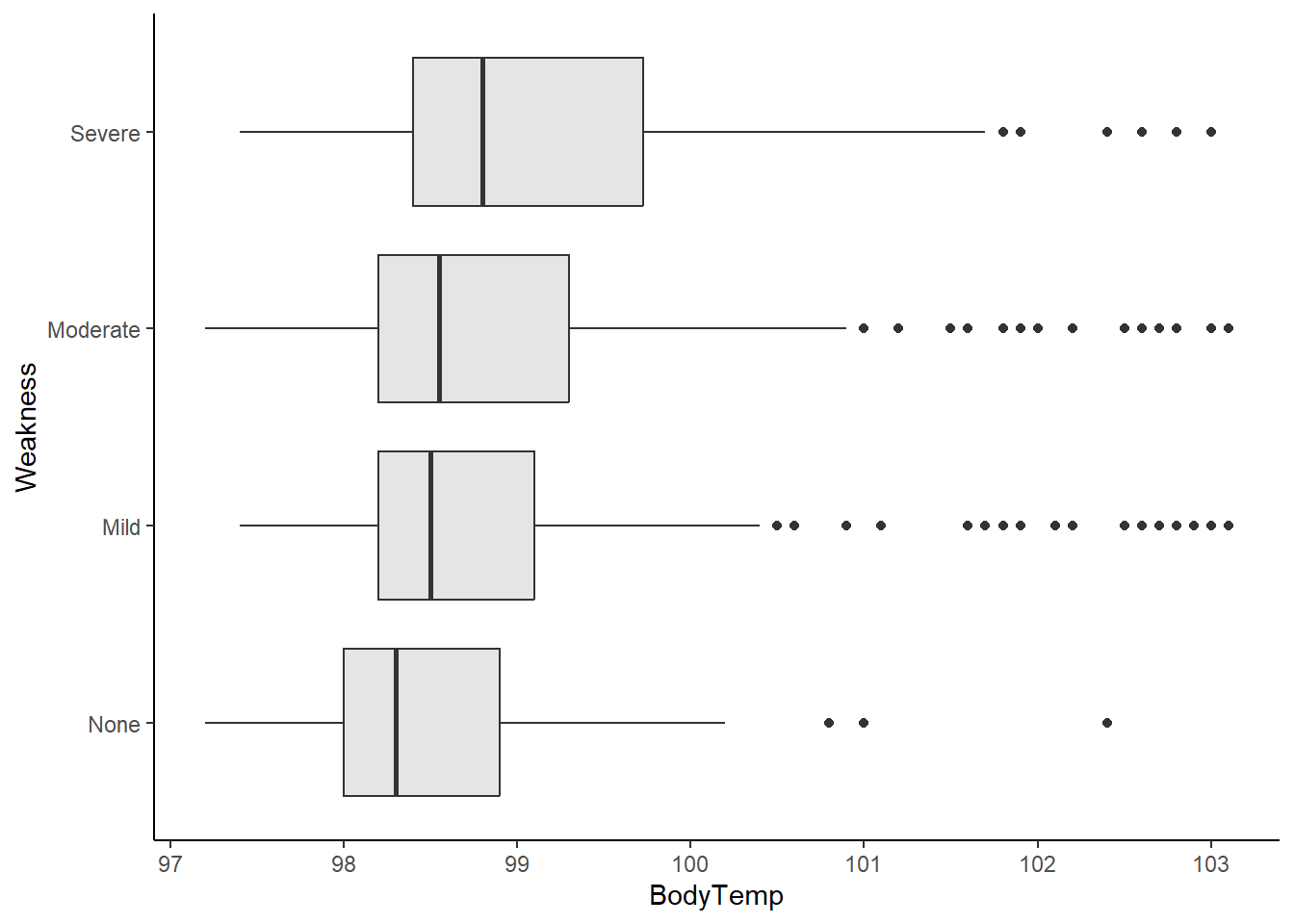
In general, higher body temperatures were recorded in patients experiencing severe weakness than those with moderate, mild, or no weakness.
Boxplot of body temperature vs cough intensity (ranked).
ggplot(flu_data, aes(BodyTemp, CoughIntensity))+
geom_boxplot(fill="gray90")+
theme_classic()
In general, higher body temperatures were recorded in patients experiencing severe cough intensity than those with moderate, mild, or no coughing.
Boxplot of body temperature vs myalgia.
ggplot(flu_data, aes(BodyTemp, Myalgia))+
geom_boxplot(fill="gray90")+
theme_classic()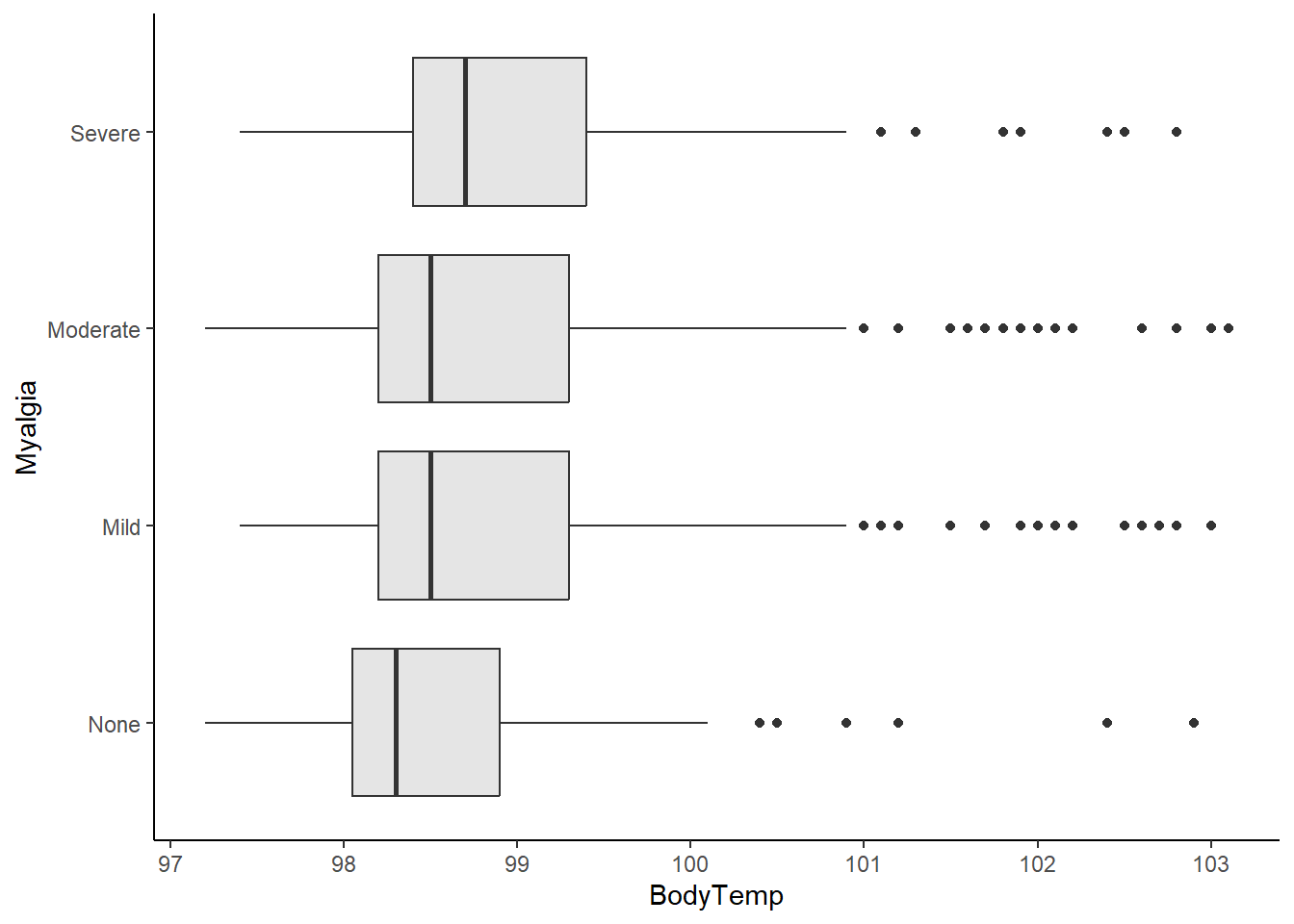
In general, higher body temperatures were recorded in patients experiencing severe myalgia than those with moderate, mild, or no myalgia.
Nausea
Now I want to visualize nausea as predicted by a number of categorical variables.
Nausea vs reported abdominal pain
ggplot(flu_data, aes(Nausea))+
geom_histogram(aes(fill=AbPain), stat = "count")+
theme_classic()Warning in geom_histogram(aes(fill = AbPain), stat = "count"): Ignoring unknown
parameters: `binwidth`, `bins`, and `pad`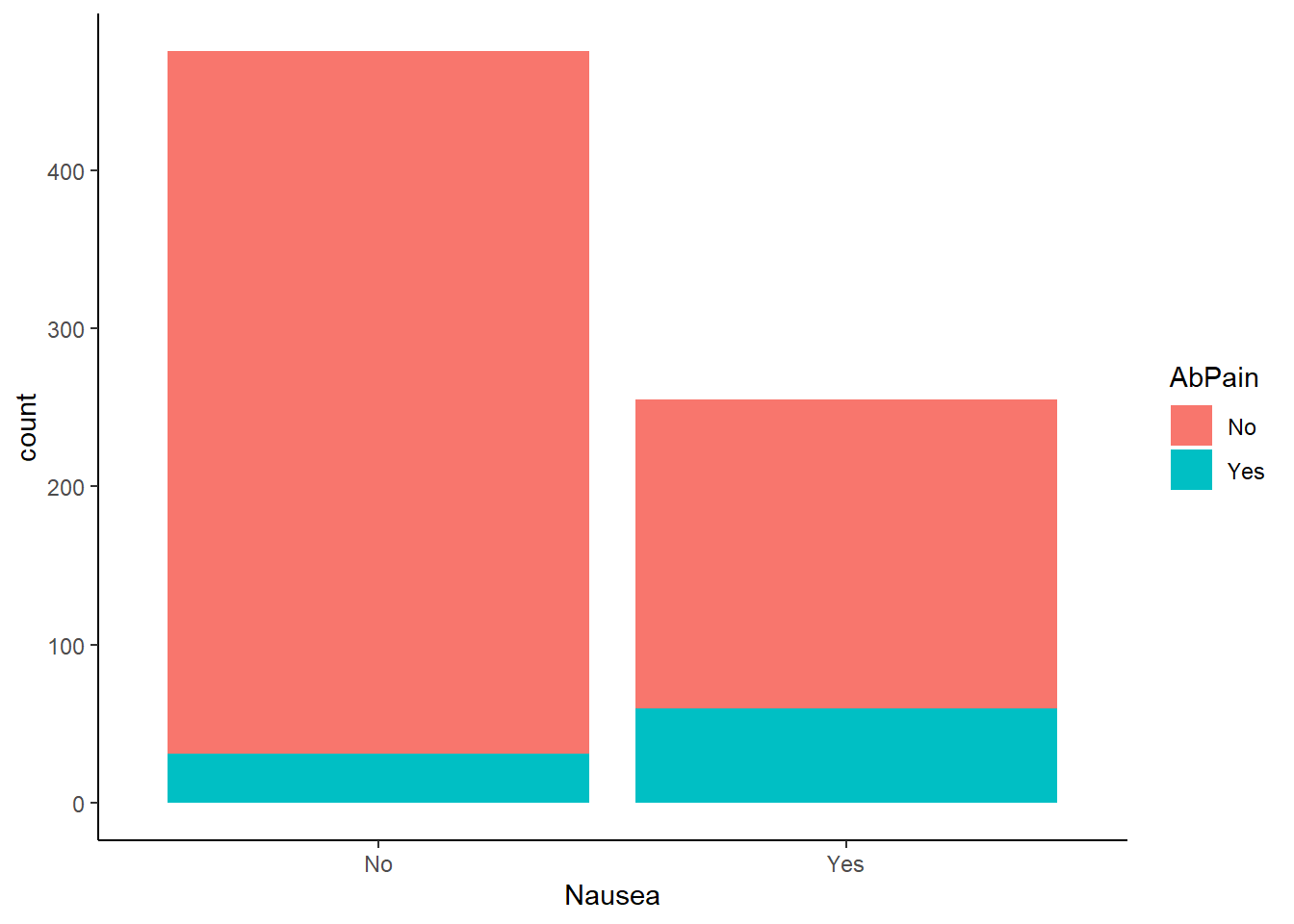
Abdominal pain was reported more often in patients experiencing nausea.
Nausea vs reported vomitting
ggplot(flu_data, aes(Nausea))+
geom_histogram(aes(fill=Vomit), stat = "count")+
theme_classic()Warning in geom_histogram(aes(fill = Vomit), stat = "count"): Ignoring unknown
parameters: `binwidth`, `bins`, and `pad`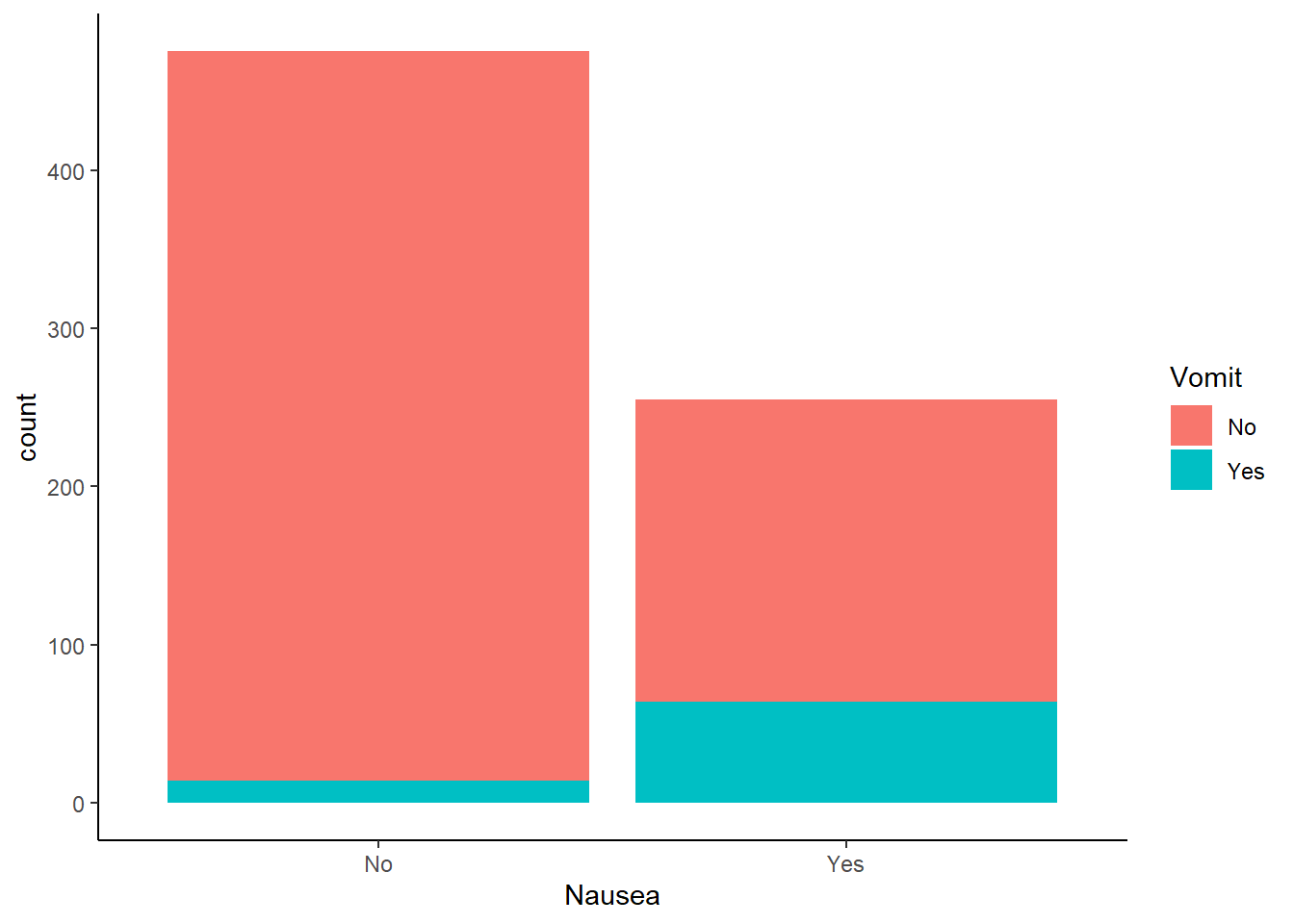
Vomitting was reported more often in patients experiencing nausea.
Nausea vs reported diarrhea
ggplot(flu_data, aes(Nausea))+
geom_histogram(aes(fill=Diarrhea), stat = "count")+
theme_classic()Warning in geom_histogram(aes(fill = Diarrhea), stat = "count"): Ignoring
unknown parameters: `binwidth`, `bins`, and `pad`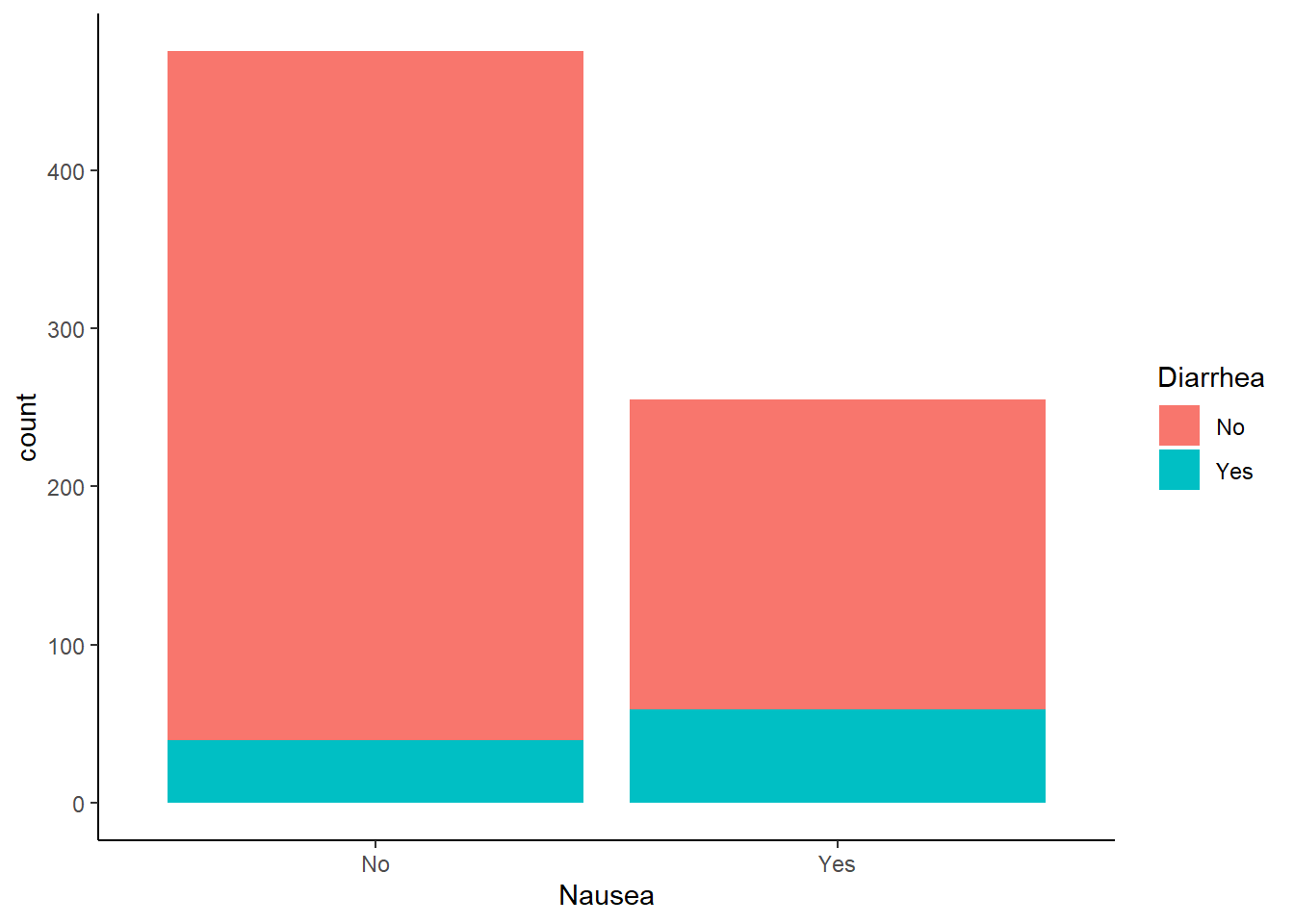
Diarrhea was reported more often in patients experiencing nausea.
Nausea vs reported chills/sweats
ggplot(flu_data, aes(Nausea))+
geom_histogram(aes(fill=ChillsSweats), stat = "count")+
theme_classic()Warning in geom_histogram(aes(fill = ChillsSweats), stat = "count"): Ignoring
unknown parameters: `binwidth`, `bins`, and `pad`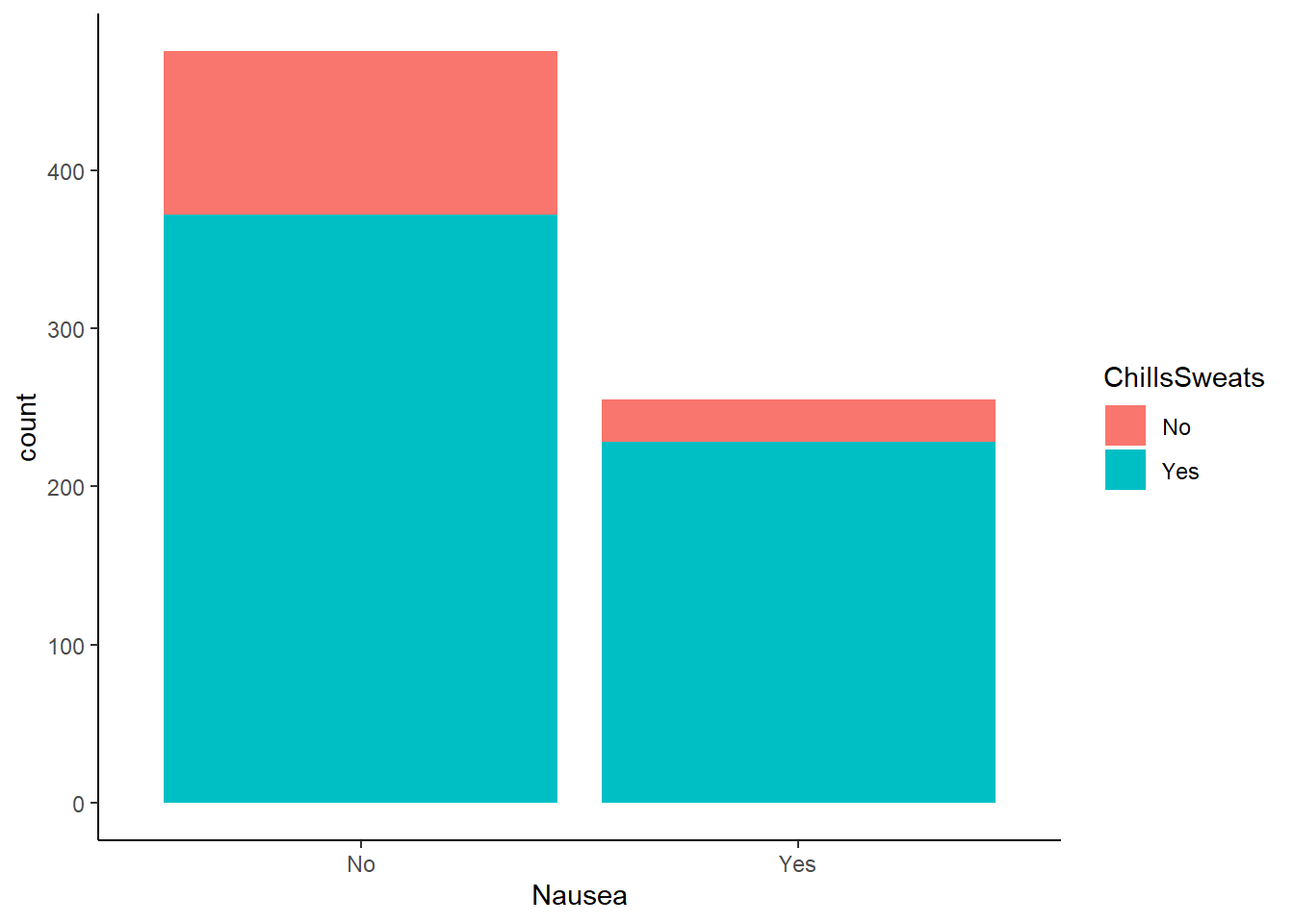
Chills/sweats were reported more often in patients experiencing nausea.